Collaborative research carried out by researchers from the School of Medicine, Tsinghua University, and Cambridge University, United Kingdom, set up machine learning-assisted high-content image analysis to study pluripotent stem cell-derived embryos in vitro. The study enabled automated and high-throughput analysis of stem cell-based embryo models.
Stem cell-based embryo models synthesized by laboratory cultured pluripotent and extraembryonic lineage stem cells are novel platforms to model mammalian embryo development. Previously, analyses of stem cell-based embryo models were mainly done through researchers’ visual inspection. The drawback is low efficiency, lack of standardization, and bias from different researchers. Moreover, it is hard to determine the variation between different pluripotent stem cell (PSC) lines due to their genetic background, derivation, culture, and reprogramming methods. Therefore, an automated, high-throughput, multi-dimensional, and unbiased workflow to quantitatively analyze stem cell-based embryo models is needed.
To facilitate the efficient and unbiased analysis of the stem cell-based embryo model, the team set up a workflow to use machine learning-assisted high-content analysis to study embryo-like structures derived from several induced pluripotent stem cell (iPSC) lines and embryonic stem cell (ESC) lines. Moreover, using this system, the team screened 55 small molecules(https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/small-molecule) and cytokines and found that BMP4 is the best candidate to facilitate the formation and the development of the iPSC/ESC and trophectoderm stem cell assembled embryos termed ITS or ETS embryos.
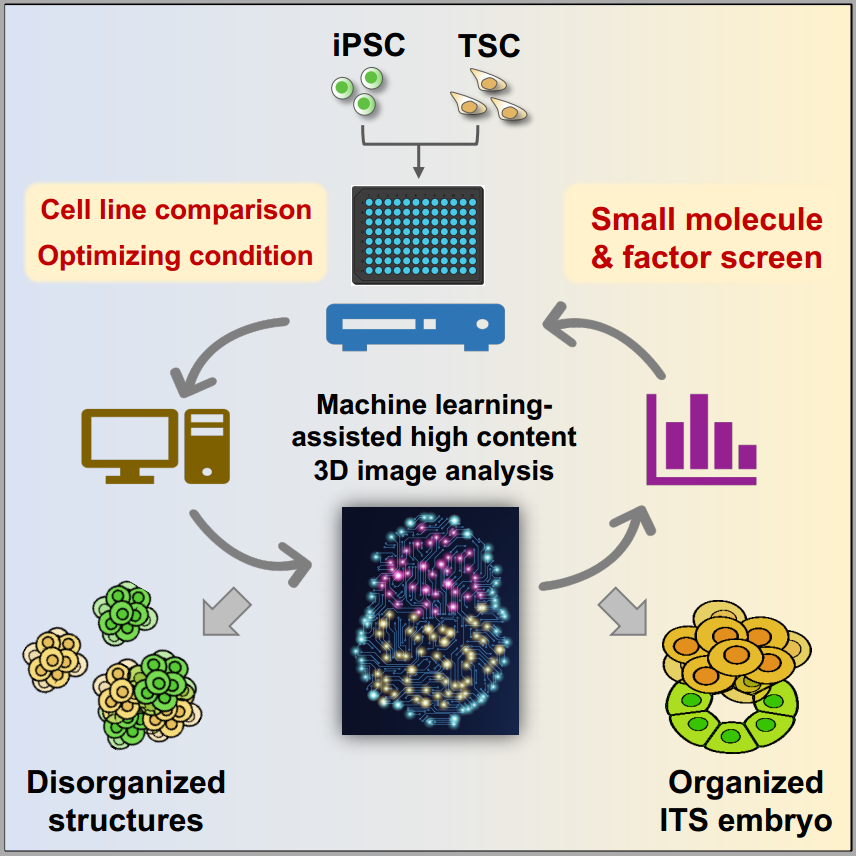
Machine learning assisted analyzing strategy for stem cell-based embryo
In the study, the researchers first set up a 3D condition to co-culture iPSCs and extraembryonic stem cells to form embryo-like structures, termed ITS, mimicking the early developmental events including pre-amonic cavity formation, basement membrane assembly, symmetry breaking, and mesoderm induction.
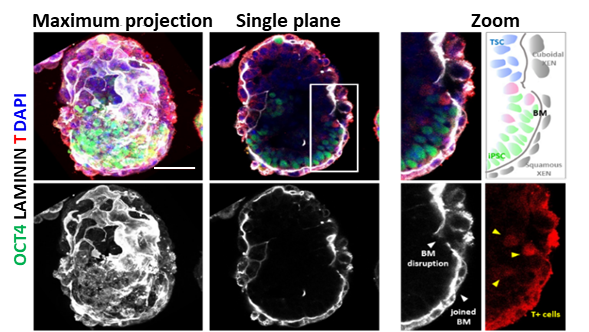
Stem cell-based embryo model formed by mouse iPSC and extraembryonic stem cell
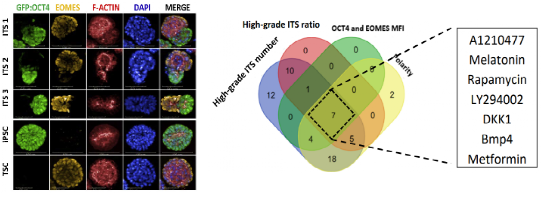
Next, the researchers set up a machine learning workflow to extract multi-dimensional features and quantify ITS embryos using 3D images collected from a high-content screening (HCS) system. The machine learning protocol automatically scanned tens of thousands of pictures and extracted hundreds of multivariate features from each ITS embryos. Then it could recognize high-grade ITS examples from the whole culture with an Accuracy of 0.92 and F1 score of 0.80. The machine learning protocol was able to calculate complex morphology features, polarization, the ratio of high-grade ITS, the quality of embryonic and extraembryonic parts of ITS embryos.
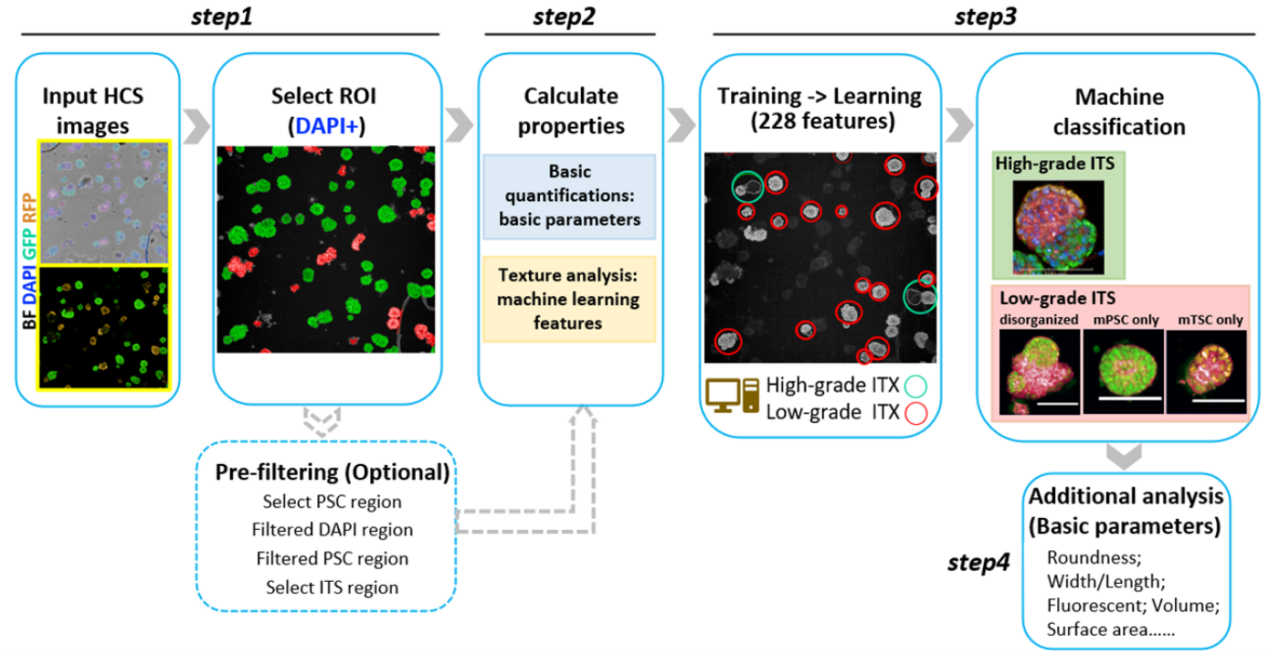
Detailed machine learning protocol for analyzing stem cell-based embryos
Using the machine learning protocol, researchers compared different PSC lines’ ability to form embryo-like structures and conducted a chemical molecule and cytokine screen. They identified that BMP4 best promoted the morphogenesis of the ITS embryo and explored the molecular mechanism through transcriptomic and epigenetic analysis.
High-content screening of small molecules and cytokines identified BMP4 as a potent inducer of ITS embryo morphogenesis
In summary, the team established robust, unbiased, and automated machine learning-based protocols to evaluate multiple iPSC and ESC lines and perform cytokine and small-molecule screens to find factors that can promote ITS embryo generation. The study revealed the variation among different iPSC and ESC lines in forming embryo-like structures. These approaches and findings are highly informative and valuable for future studies of stem cell-based embryo and organ models.
The results of this study were published online on April 23, 2021, under the title “Machine Learning-Assisted High-Content Analysis of Pluripotent Stem Cell-Derived Embryos in vitro” inStem Cell Reports, a leading stem cell biology journal.
Reference:
Jianying Guo,1,2,6Peizhe Wang,1,6Berna Sozen,3,4Hui Qiu,1,2Yonglin Zhu,1Xingwu Zhang,1Jia Ming,1Magdalena Zernicka-Goetz,3,5,*and Jie Na1,*Machine Learning-Assisted High-Content Analysis of Pluripotent Stem Cell-Derived Embryos in vitro.Stem Cell Reports,2021,16:1–16
Link to full article:
https://www.cell.com/stem-cell-reports/fulltext/S2213-6711(21)00148-X